In [1]:
import pandas as pd, seaborn as sns, numpy as np
sns.set(font='Bitstream Vera Sans')
sns.set_context('poster', {'figsize':(12,8)})
pd.options.display.max_rows = 8
%matplotlib inline
%cd ~/2014_fall_ASTR599/notebooks/
%run talktools.py
%cd ~/notebook/
[Errno 2] No such file or directory: '/Users/jakevdp/2014_fall_ASTR599/notebooks/' /Users/jakevdp/Opensource/2014_fall_ASTR599/notebooks
[Errno 2] No such file or directory: '/Users/jakevdp/notebook/' /Users/jakevdp/Opensource/2014_fall_ASTR599/notebooks

This notebook was put together by Abraham Flaxman for UW's Astro 599 course. Source and license info is on GitHub.

In [2]:
import pandas as pd
big data?¶
medium data¶
learning objectives:¶
- know that
pandasexists --- tool for manipulating medium sized tabular data - know how to learn more about
pandas - get some hands on experience with the
pandas.DataFramefor data manipulation
pandas exists¶

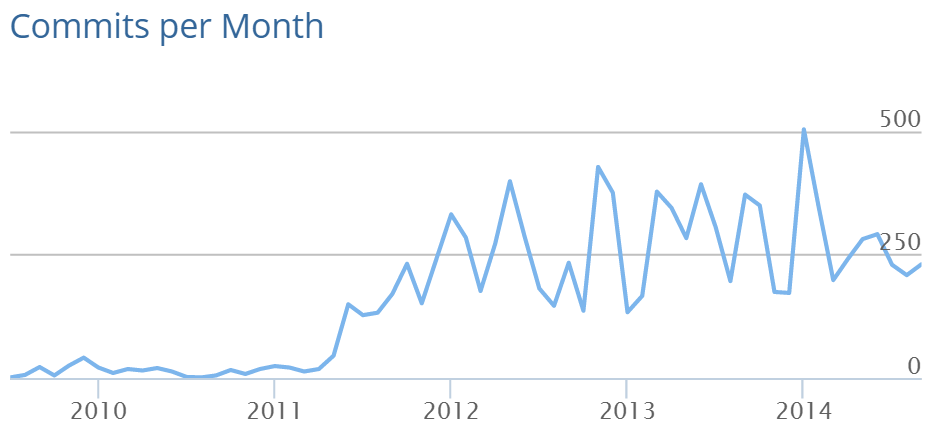
In [3]:
import pandas as pd # we did this already!
# did it work?
In [4]:
pd.__version__
Out[4]:
'0.14.1'
how to learn more¶

In [ ]:
pd.DataFrame([shift-tab]
hands on experience¶
- making
DataFrames - using tabular data
- reshaping tables
- combining tables
- grouping, mapping, and applying
- dealing with date and time data
making DataFrames¶
In [6]:
# loading a csv
df = pd.read_csv(fname)
--------------------------------------------------------------------------- NameError Traceback (most recent call last) <ipython-input-6-30775157dcae> in <module>() 1 # loading a csv ----> 2 df = pd.read_csv(fname) NameError: name 'fname' is not defined
In [ ]:
# loading an excel file
df = pd.read_[tab]
In [ ]:
# loading a stata file
df = pd.read_
making DataFrames from scratch¶
In [7]:
df = pd.DataFrame({'a': [10,20,30],
'b': [40,50,60]})
In [8]:
df
Out[8]:
| a | b | |
|---|---|---|
| 0 | 10 | 40 |
| 1 | 20 | 50 |
| 2 | 30 | 60 |
In [18]:
# think of DataFrames as numpy arrays plus
df.columns
df.index
df.b.index is df.index
Out[18]:
True
questions?¶
aside: global health¶

our running example¶
From Global Health Data Exchange, load PHMRC VA adult data, CSV format:
In [19]:
url = 'http://ghdx.healthdata.org/sites/default/files/'\
'record-attached-files/IHME_PHMRC_VA_DATA_ADULT_Y2013M09D11_0.csv'
In [20]:
df = pd.read_csv(url)
/Users/jakevdp/anaconda/lib/python2.7/site-packages/pandas/io/parsers.py:1139: DtypeWarning: Columns (18,29,38,41,60,96) have mixed types. Specify dtype option on import or set low_memory=False. data = self._reader.read(nrows)
In [21]:
df = pd.read_csv(url, low_memory=False)
what's in df?¶
In [22]:
df
Out[22]:
| site | module | gs_code34 | gs_text34 | va34 | gs_code46 | gs_text46 | va46 | gs_code55 | gs_text55 | ... | word_woman | word_womb | word_worri | word_wors | word_worsen | word_worst | word_wound | word_xray | word_yellow | newid | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Mexico | Adult | K71 | Cirrhosis | 6 | K71 | Cirrhosis | 8 | K71 | Cirrhosis | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 1 | AP | Adult | G40 | Epilepsy | 12 | G40 | Epilepsy | 16 | G40 | Epilepsy | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 2 | AP | Adult | J12 | Pneumonia | 26 | J12 | Pneumonia | 37 | J12 | Pneumonia | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| 3 | Mexico | Adult | J33 | COPD | 8 | J33 | COPD | 10 | J33 | COPD | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 7837 | Dar | Adult | ZZ23 | Other Cardiovascular Diseases | 22 | ZZ23 | Other Cardiovascular Diseases | 31 | ZZ23 | Other Cardiovascular Diseases | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7843 |
| 7838 | AP | Adult | T36 | Poisonings | 27 | T36 | Poisonings | 38 | T36 | Poisonings | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7844 |
| 7839 | UP | Adult | X09 | Fires | 15 | X09 | Fires | 19 | X09 | Fires | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7845 |
| 7840 | Dar | Adult | C15 | Esophageal Cancer | 13 | C15 | Esophageal Cancer | 17 | C15 | Esophageal Cancer | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7846 |
7841 rows × 946 columns
In [23]:
# also load codebook (excel doc)
url = 'http://ghdx.healthdata.org/sites/default/files/'\
'record-attached-files/IHME_PHMRC_VA_DATA_CODEBOOK_Y2013M09D11_0.xlsx'
cb = pd.read_excel(url)
In [27]:
cb.head()
Out[27]:
| variable | question | module | health_care_experience | coding | |
|---|---|---|---|---|---|
| 0 | site | Site | General | 0 | NaN |
| 1 | newid | Study ID | General | 0 | NaN |
| 2 | gs_diagnosis | Gold Standard Diagnosis Code | General | 0 | NaN |
| 3 | gs_comorbid1 | Gold Standard Comorbid Conditions 1 | General | 0 | NaN |
| 4 | gs_comorbid2 | Gold Standard Comorbid Conditions 2 | General | 0 | NaN |
using tabular data¶
In [29]:
# each column of pd.DataFrame is a pd.Series
cb.module
Out[29]:
0 General 1 General ... 1647 Neonate 1648 Neonate Name: module, Length: 1649, dtype: object
In [44]:
# can uses square-brackets instead of "dot"
# (useful if column name has spaces!)
cb.iloc[3, 3]
Out[44]:
0
In [45]:
# what's in this series?
cb.module.value_counts()
Out[45]:
Adult 902 Child 466 Neonate 218 General 63 dtype: int64
In [46]:
# accessing individual cells
df.head()
Out[46]:
| site | module | gs_code34 | gs_text34 | va34 | gs_code46 | gs_text46 | va46 | gs_code55 | gs_text55 | ... | word_woman | word_womb | word_worri | word_wors | word_worsen | word_worst | word_wound | word_xray | word_yellow | newid | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Mexico | Adult | K71 | Cirrhosis | 6 | K71 | Cirrhosis | 8 | K71 | Cirrhosis | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 1 | AP | Adult | G40 | Epilepsy | 12 | G40 | Epilepsy | 16 | G40 | Epilepsy | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 2 | AP | Adult | J12 | Pneumonia | 26 | J12 | Pneumonia | 37 | J12 | Pneumonia | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| 3 | Mexico | Adult | J33 | COPD | 8 | J33 | COPD | 10 | J33 | COPD | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| 4 | UP | Adult | I21 | Acute Myocardial Infarction | 17 | I21 | Acute Myocardial Infarction | 23 | I21 | Acute Myocardial Infarction | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
5 rows × 946 columns
In [47]:
# to access by row and column
# can use names
df.loc[4, 'gs_text34']
Out[47]:
'Acute Myocardial Infarction'
In [48]:
# or use numbers
df.iloc[4, 3]
Out[48]:
'Acute Myocardial Infarction'
In [49]:
# same because columns 3 is gs_text
# how to check?
df.columns[3]
Out[49]:
'gs_text34'
improve this example¶
In [50]:
df.head()
Out[50]:
| site | module | gs_code34 | gs_text34 | va34 | gs_code46 | gs_text46 | va46 | gs_code55 | gs_text55 | ... | word_woman | word_womb | word_worri | word_wors | word_worsen | word_worst | word_wound | word_xray | word_yellow | newid | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Mexico | Adult | K71 | Cirrhosis | 6 | K71 | Cirrhosis | 8 | K71 | Cirrhosis | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 1 | AP | Adult | G40 | Epilepsy | 12 | G40 | Epilepsy | 16 | G40 | Epilepsy | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 2 | AP | Adult | J12 | Pneumonia | 26 | J12 | Pneumonia | 37 | J12 | Pneumonia | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| 3 | Mexico | Adult | J33 | COPD | 8 | J33 | COPD | 10 | J33 | COPD | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| 4 | UP | Adult | I21 | Acute Myocardial Infarction | 17 | I21 | Acute Myocardial Infarction | 23 | I21 | Acute Myocardial Infarction | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
5 rows × 946 columns
In [51]:
# make the row names more interesting than numbers starting from zero
df.index = ['person %d'%(i+1) for i in df.index]
In [52]:
# have a look at the first ten rows and first five columns
df.iloc[:10, :5]
Out[52]:
| site | module | gs_code34 | gs_text34 | va34 | |
|---|---|---|---|---|---|
| person 1 | Mexico | Adult | K71 | Cirrhosis | 6 |
| person 2 | AP | Adult | G40 | Epilepsy | 12 |
| person 3 | AP | Adult | J12 | Pneumonia | 26 |
| person 4 | Mexico | Adult | J33 | COPD | 8 |
| ... | ... | ... | ... | ... | ... |
| person 7 | Dar | Adult | N17 | Renal Failure | 29 |
| person 8 | Dar | Adult | B20 | AIDS | 1 |
| person 9 | Bohol | Adult | C34 | Lung Cancer | 19 |
| person 10 | UP | Adult | O67 | Maternal | 21 |
10 rows × 5 columns
In [53]:
# slicing with named indices is possible, too
df.loc['person 10', 'site':'gs_text34']
Out[53]:
site UP module Adult gs_code34 O67 gs_text34 Maternal Name: person 10, dtype: object
notice anything weird about that, though?
what else can we do?¶
In [54]:
# logical operations (element-wise)
df.module == "Adult"
Out[54]:
person 1 True person 2 True ... person 7840 True person 7841 True Name: module, Length: 7841, dtype: bool
In [55]:
# usefule for selecting subset of rows
dfa = df[df.module == "Adult"]
In [56]:
# summarize
dfa.va34.describe(percentiles=[.025,.975])
Out[56]:
count 7841.000000 mean 18.998342 std 9.929374 min 1.000000 2.5% 1.000000 50% 21.000000 97.5% 34.000000 max 34.000000 dtype: float64
In [57]:
# percent of occurrences
100 * dfa.gs_text34.value_counts(normalize=True).round(3)
Out[57]:
Stroke 8.0 Other Non-communicable Diseases 7.6 ... Asthma 0.6 Esophageal Cancer 0.5 Length: 34, dtype: float64
In [58]:
# calculate average
dfa.word_fever.mean()
Out[58]:
0.18339497513072311
In [59]:
# create visual summaries
dfa.site.value_counts(ascending=True).plot(kind='barh', figsize=(12,8))
Out[59]:
<matplotlib.axes._subplots.AxesSubplot at 0x10a56eb50>
pandas.DataFrame.GroupBy¶
In [60]:
df.groupby('gs_text34')
Out[60]:
<pandas.core.groupby.DataFrameGroupBy object at 0x10a55d310>
In [ ]:
df.word_activ
In [61]:
df.groupby('gs_text34').word_fever.mean()
Out[61]:
gs_text34 AIDS 0.284861 Acute Myocardial Infarction 0.135000 ... Suicide 0.032258 TB 0.192029 Name: word_fever, Length: 34, dtype: float64
In [64]:
(dfa.groupby('gs_text34').word_activ.mean() * 100).order().plot(kind='barh', figsize=(12,12))
Out[64]:
<matplotlib.axes._subplots.AxesSubplot at 0x10bcce3d0>
reshaping tables¶
In [63]:
df.filter(like='word').head()
Out[63]:
| word_abdomen | word_abl | word_accid | word_accord | word_ach | word_acidosi | word_acquir | word_activ | word_acut | word_add | ... | word_wit | word_woman | word_womb | word_worri | word_wors | word_worsen | word_worst | word_wound | word_xray | word_yellow | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| person 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| person 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| person 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| person 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| person 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
5 rows × 679 columns
In [ ]:
# get a smaller table to experiment with
t = df.filter(like='word').describe()
t
In [ ]:
# transpose table
t.T.head()
In [ ]:
# convert from "wide" to "long"
t_long = t.T.stack()
t_long
In [ ]:
# convert back
t_long.unstack()
In [ ]:
# challenge: find the most common word
pandas.pivot_table¶
In [ ]:
df.head()
In [ ]:
piv = pd.pivot_table(df, values='word_fever',
index=['site'], columns=['gs_text34'],
aggfunc=sum)
piv
inverse-pivot-tables: pandas.melt¶
In [ ]:
causes = list(piv.columns)
piv['site'] = piv.index
fever_cnts = pd.melt(piv, id_vars=['site'], value_vars=causes,
var_name='cause of death',
value_name='number w fever mentioned')
fever_cnts.head()
In [ ]:
# cleaner to drop the rows with NaNs
fever_cnts.dropna().head()
appending DataFrames¶
In [ ]:
# put together first three and last three rows of df
df.iloc[:3].append(df.iloc[-3:])
merging DataFrames¶
In [ ]:
df.iloc[:5,20:25]
In [ ]:
cb.iloc[10:15]
merge question text into df¶
In [ ]:
pd.merge(
In [ ]:
merged_df = pd.merge(df.T, cb, left_index=True, right_on='variable')
merged_df.filter(['variable', 'question', 'person 1', 'person 2']).dropna().head(10)
GroupBy¶
In [ ]:
df.groupby('gs_text34').word_fever.mean()
In [ ]:
for g, dfg in df.groupby('gs_text34'):
print g
# process DataFrame for this group
break
In [ ]:
# apply
def my_func(row):
return row['word_xray'] > 0
my_func(df.iloc[0])
df.apply(my_func, axis=1)
In [ ]:
rng = pd.date_range('1/1/2012', periods=1000, freq='S')
In [ ]:
rng
In [ ]:
ts = pd.Series(np.random.randint(0, 500, len(rng)), index=rng)
In [ ]:
ts.resample('5Min', how='sum')
Exercise¶
Use the codebook to find the column which corresponds to the question "Did ____ have a fever?"¶
In [ ]:
# in here:
cb.head()
In [ ]:
# hint: look here
cb.question.str
In [ ]:
var =
The "endorsement rate" for a sign or symptom of disease is the fraction of verbal autopsy interviews where the respondent answered "Yes" to the question about that sign/symptom.
Find the cause with the highest endorsement rate for the symptom "fever" among Adult deaths¶
In [ ]:
# create a new column with 0/1 values instead of Yes/No/Don't Know/Refused to Answer
df['fever'] = df[var] == 'Yes'
In [ ]:
# use groupby like we did in class
Display the cause-specific endorsement rates visually¶
In [ ]:
import matplotlib.pyplot as plt
# do some plotting like we did in class
plt.xlabel('Endorsement Rate (%)')
Find the question that asks if the deceased had AIDS¶
In [ ]:
# this might be familiar now...
Make a 2x2 table showing the number of adult deceased with and without this question endorsed for decedents with underlying cause AIDS and not AIDS¶
In [ ]:
# could be good to start by making some numeric columns like above
In [ ]:
# groupby is a way to go, but not the only way
Compare this 2x2 table across study sites¶
In [ ]:
# note: there are lots of ways to do this
# try to find one that is so simple, you will understand it next time you look
Now do a series of 2x2 tables comparing the percent of deaths truely due to AIDS for which "Had AIDS" question was endorsed¶
In [ ]:
# some interesting differences. if you want to know why, you might be a social scientist...